Oligo Pools
Cost effective high-throughput DNA Synthesis
Nucleic acid synthesis is the backbone of innovation in life sciences—powering breakthroughs across biomedicine, genomics, synthetic biology, and beyond. As the demand for faster, more scalable, and cost-effective solutions grows, ultra-high-throughput nucleic acid synthesis has emerged as a game-changing technology with transformative potential across industries. At Watsonbio, we are redefining what’s possible in next-generation nucleic acid synthesis. We’ve broken through the limitations of traditional methods with our proprietary DYHOW platform—a next-generation, ultra-high-throughput DNA synthesis system built from the ground up with independent intellectual property.
Our latest innovation, DYHOW-3B, delivers unprecedented performance:
- Synthesis of up to 4.35 million unique custom DNA sequences
- Maximum oligo length of 230 nucleotides
- Remarkably high accuracy and uniformity
- Significantly reduced synthesis costs and turnaround time
With DYHOW-3B, Watsonbio empowers rapid innovation across synthetic biology, medicine, agriculture, food science, and more—solving real-world challenges from research to industrial production.
Workflows

Highly Uniform and Accurate Synthesis
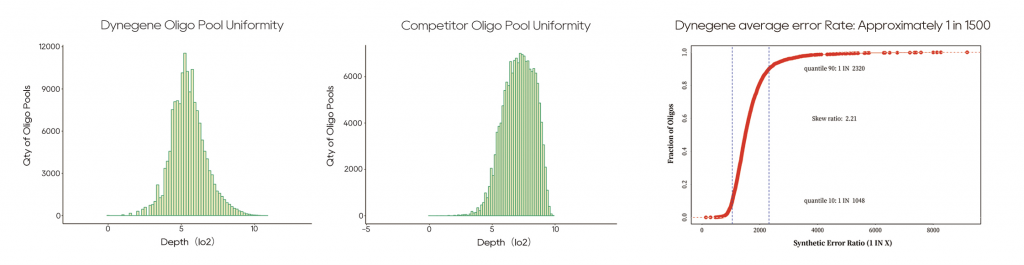
NGS sequencing verification of oligo pools with identical sequences synthesized separately by Watsonbio and a competitor revealed superior uniformity in Watsonbio’s synthesized oligo pools.
Application Example
Oligo pool synthesis is the foundation of library construction. To build a successful library, strict control is necessary throughout the synthesis process. Leveraging the high-throughput DNA synthesis platform developed by Watsonbio, we constructed a library containing 63,950 sgRNA sequences. NGS verification demonstrated high coverage and uniformity of the sgRNA library.
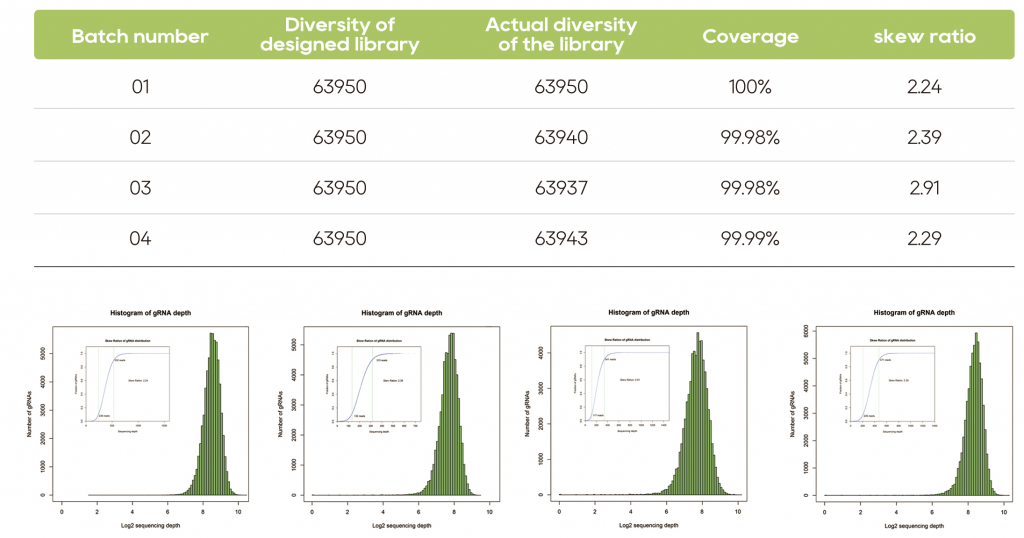
Analysis of NGS data from four batches of oligo pool-constructed libraries revealed high coverage, uniform distribution, and excellent stability across batches of the sgRNA libraries.
