Multiplex miRNA Assay
SimPlex-miR™ enables fast, accurate miRNA profiling in 20 minutes—one tube, no PCR, no compromise.
SimPlex-miR™ : Simple multiplex miRNA
-
Innovation: Multiple miRNAs in one tube within 20 min without PCR amplification.
-
High Fidelity miRNAs Profiles: Solve the bottle neck of miRNAs biomarker assay, including batch variations of isolation yield, bias of miRNA labelling/ligation, and bias of PCR amplification from GC contents and annealing temperature.
-
Applications: Liquid biopsy of cancer early diagnostics, drug selection and R&D for new therapeutics.
SimPlex-miR™ overcomes the bias from PCR amplification (Company Q) and redundant operations and long/high temperature hybridization (Company T)
Challenges - Limitations of Current PCR Based miRNA Assay
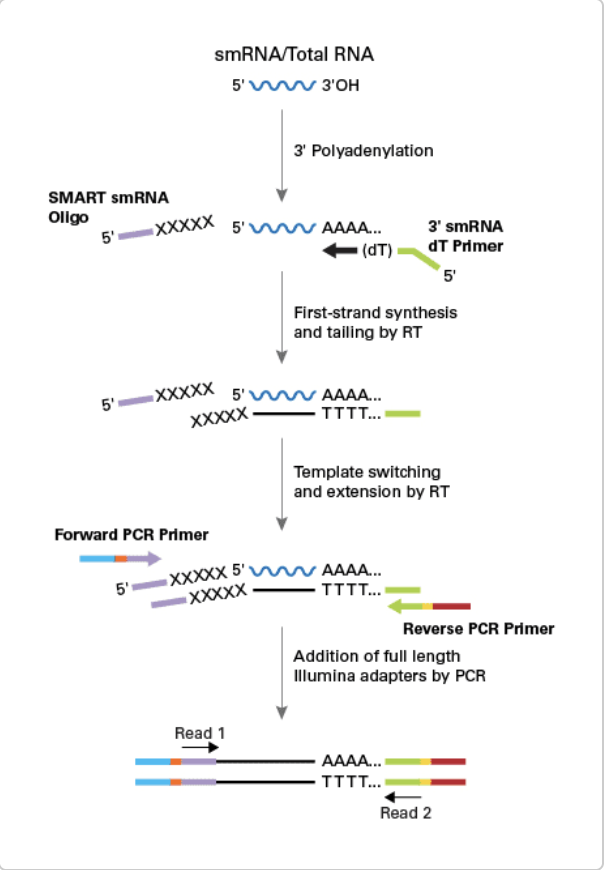
Mass of total miRNAs is less than 0.1% from sera, and consists of 13,000 kinds of different sequences.

- Batch Variations of miRNA Isolation
- Consistency from different measurement and different group
- Biases in RNA Ligation & PCR Amplification
- Significant biases introduced by RNA ligation lead to inaccurate miRNA quantification by 1000 folds.
- Respective miRNA G/C content greatly influences rates of cDNA synthesis and is also responsible for template-specific preferences in PCR amplification.
Raabe et al., Nucleic Acids Research, 2014
Innovations - SimPlex-miR™ Providing High Fidelity Profiles
Accuracy from SimPlex: 20 min. mix/hybridization + 1 min analysis.
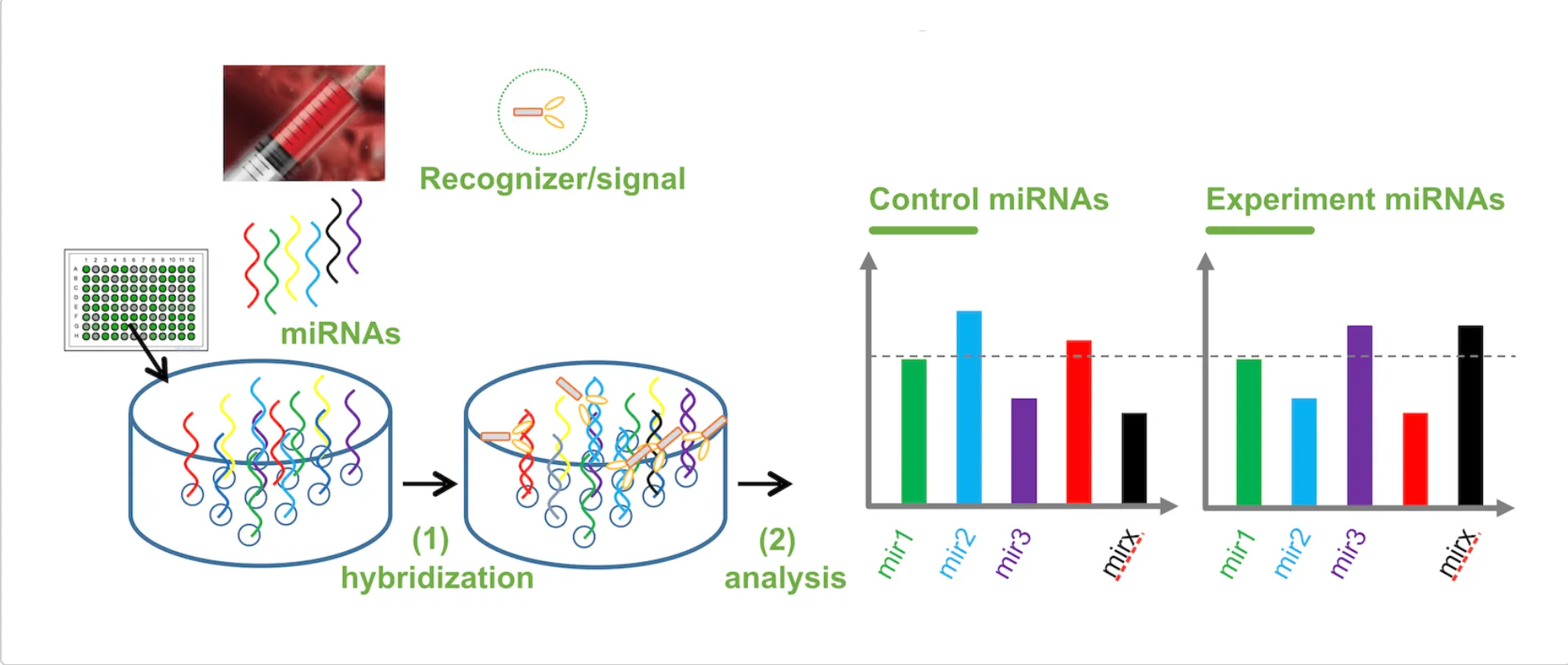
- Short hybridization time reduces the possibility of miRNA degradation and non-specific adsorption.
- Multiplex miRNA assay and analysis methodology solves the batch variations.
Summary of SimPlex-miR™
Applicable to other type of linear RNA, such as IncRNA, mRNA
Cost reduced to 5-10% of other miRNAs assay
High Fidelity miRNAs Profiling
- No PCR bias
- No miRNA ligation bias
- No miRNA isolation batch variations
- Short assay time, less miRNA degradation and nonspecific adsorption
Please contact info@watsonbio.com for details
Send your request to
info@watsonbio.com
or click Request Quote below